Biotechnology : Principles and Processes: Class 12 Biology Chapter 11
Key Features of NCERT Material for Class 12 Biology Chapter 11 – Biotechnology: Principles and Processes
In the previous chapter 10 of NCERT class 12 biology: Microbes in human welfare, you studied microbes and their application in the welfare of human lives. In this chapter: Biotechnology: Principles and Processes, you will understand biotechnology and different processes associated with it.
What strikes a chord when you hear the term ‘Biotechnology’? Is it the way toward making curd or bread using microorganisms? Or on the other hand is it the way toward using gene treatment to address a damaged gene? You will be shocked to realize that both the above procedures fall under biotechnology! We should become familiar with the various apparatuses of biotechnology.
Standards Of Biotechnology
The field of biotechnology has developed immensely throughout the years. It has gone from a conventional perspective on using microorganisms to generating items for human use. The advanced perspective on using genes for creating antibodies. Hence, the European Federation of Biotechnology (EFB) has thought of another definition that includes both the conventional and the cutting edge perspective on biotechnology. It expresses that –
‘Biotechnology is the coordination of normal science and life forms, cells, parts thereof, and atomic analogues for items and administrations’. The two centre methods liable for the introduction of current biotechnology are:
Genetic Engineering: It includes strategies that change the science of the genetic material with the end goal that when they are brought into the host, the host phenotype changes.
Upkeep of a defilement free condition to empower the development of just the ideal organism in enormous amounts during the assembling of items like immunizations, anti-infection agents, and so forth.
Quick revisions notes
The methods of using live living beings or chemicals from living beings to create items and procedures valuable to people. Numerous procedures like in vitro treatment prompting ‘test-tube’ infant, orchestrating gene and using it, building up a DNA immunization or remedying a deficient gene are likewise parts of Biotechnology.
The European Federation of Biotechnology (EFB) has given a meaning of biotechnology that involves both conventional and present-day atomic biotechnology.The definition is as follow-“The joining of characteristic science and life forms, cells, parts thereof, and sub-atomic comparable to for items and administrations”.
Standards of Biotechnology
Present day biotechnology depends on two primary standards
- Genetic Engineering – Genetic Engineering is characterized as the immediate control of genome (DNA and RNA) of a creature. It includes the exchange of new genes to improve the capacity or characteristic into host life forms and in this manner changes the phenotype of the host living being.
- Maintenance of sterile condition in substance building procedure to empower development of just wanted organisms for assembling of biotechnological items like anti-toxins, antibody, chemicals and so forth.
- Traditional hybridization utilized in plants and creature reproducing prompts incorporation and increase of unwanted genes alongside the ideal attributes. The method of genetic building which incorporate production of recombinant DNA, utilization of gene cloning and gene move permit us to confine and present just one or a lot of alluring genes without bringing bothersome genes into the objective living being.
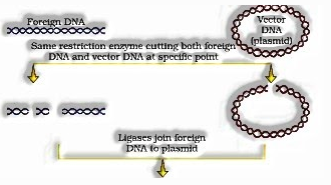
- In a chromosome there is a particular DNA sequence called the source of replication, which is liable for starting replication. So, for the increase of any outsider bit of DNA in a creature, it should be a piece of a chromosome which has a particular sequence known as ‘starting point of replication’. In this way, an outsider DNA is connected with the birthplace of replication, so that, this outsider bit of DNA can repeat and duplicate itself in the host life form. This is known as Cloning or making different indistinguishable duplicates of any layout DNA.
- The development of the main recombinant DNA rose up out of the chance of connecting a gene encoding anti-microbial obstruction with a local Plasmid of Salmonella Typhimurium.
Stanley Cohen and Herbert Boyer
Stanley Cohen and Herbert Boyer in 1972 disengaged the anti-toxin obstruction gene by removing a bit of DNA from a plasmid (independently reproducing roundabout extra-chromosomal DNA) of Salmonella typhimurium. The cutting of DNA at explicit areas got conceivable with the revelation of the supposed ‘sub-atomic scissors’– restriction chemicals.
- The cut bit of DNA was then connected with the plasmid DNA. This plasmid DNA go about as vectors to move the bit of DNA joined to it.A plasmid can be utilized as vector to convey an outsider bit of DNA into the host living being.
- The connecting of anti-infection obstruction gene with the plasmid vector become conceivable with the protein ligase, which follows up on cut DNA particles and joins their closures. This makes another mix of independently repeating DNA made in vitro and known as recombinant DNA.
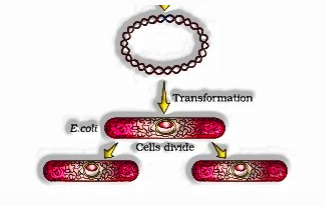
- When this DNA is moved into E.coli, it could reproduce using the new host DNA polymerase compound and make different duplicates. The capacity to increase duplicates of anti-microbial opposition gene in E.coli was called cloning of anti-microbial obstruction gene in E.coli.
“Recombinant DNA innovation” or additionally called “Genetic Engineering” bargains about, the creation of new mixes of genetic material (falsely) in the research facility. These “recombinant DNA” (rDNA) particles are then brought into host cells, where they can be spread and increased.
Steps of Fecombinant DNA Technology –
- Distinguishing proof of DNA with alluring genes.
- Presentation of the recognized DNA into the host.
III. Upkeep of presented DNA in the host and move of the DNA to its offspring.
Instruments of Recombinant DNA Technology incorporates
- Restriction Enzymes
- Polymerase proteins
- Ligases
- Vectors
- Host living beings
Restriction Enzymes (Molecular Scissors):
Restriction compounds have a place with a bigger class of catalysts called Nucleases. There are of two sorts; Exonucleases and Endonucleases. Exonucleases expel nucleotides from the closures of the DNA while, endonucleases make cuts at explicit situation inside the DNA.
Model, the primary restriction endonuclease – Hind II, consistently cut DNA particles at a specific point by perceiving a particular sequence of six base sets. This particular base sequence is known as the Recognition Sequence for Hind II.
- Each restriction endonuclease perceives a particular palindromic nucleotide sequence in the DNA. Palindromes are gathering of letters that structure similar words when perused both forward and in reverse for instance “MALYALAM”.
5′ — GAATTC — 3′
3′ — CTTAAG — 5′
The palindrome in DNA is a sequence of base combines that peruses same on two stands when the direction of perusing is kept the equivalent.
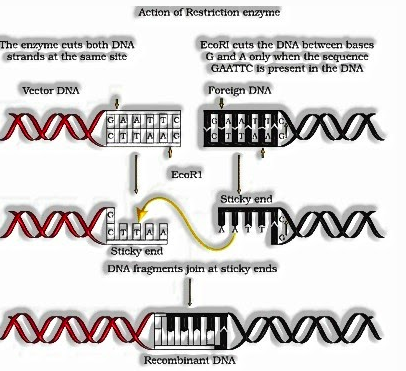
- Restriction proteins cut the strand of DNA somewhat away from the focal point of the palindrome site between a similar two bases on the contrary strands having clingy strand. The tenacity of the strands offices the activity of the catalyst DNA ligase.
- Restriction endonucleases are utilized in genetic building to shape recombinant atoms of DNA which are made out of DNA from various sources or genome.
- When cut a similar restriction catalyst the resultant DNA sections have a similar sort of Sticky-closes and can be consolidated using DNA ligases.
Diagrammatic portrayal of Recombinant DNA innovation
Detachment and seclusion of DNA pieces
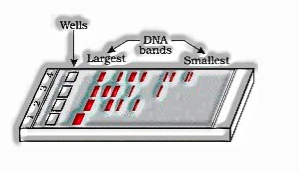
The part of DNA got by cutting DNA using restriction catalyst is isolated by procedure called gel electrophoresis. Adversely charged DNA parts can be isolated by driving them to move towards the anode under an electric field through medium. DNA parts separate as per their size through sieving impact gave by agarose gel.
- The isolated DNA piece can be imagined subsequent to recoloring the DNA with ethodium bromide followed by presentation to UV light. Isolated groups of DNA are isolated from agarose gel and separated from gel, called elution. The DNA part refined along these lines is utilized for recombination.
Cloning Vector
Plasmids and Bacteriophages is generally utilized vector for cloning. They have capacity to recreate inside bacterial cells free of the control of chromosomal DNA. Bacteriophages on account of their high number per cell, have exceptionally high duplicate quantities of their genome inside the bacterial cells.
Following highlights are required to encourage cloning into a vector-
- Root of replication (ori) – the sequence from where replication begins and any bit of DNA when connected to this sequence can be made to imitate inside the host cells.This sequence is liable for controlling the duplicate number of the connected DNA.
- Selectable marker-help in the recognizing and disposing of non transformants and specifically allowing the development of the transformants. Change is a technique through which a bit of DNA is presented in a host bacterium. Generally,the genes encoding protection from anti-infection agents, for example, ampicillin, chloramphenicol, antibiotic medication or kanamycin, and so forth., are viewed as helpful selectable markers for E. coli.
- Cloning destinations to interface the unfamiliar DNA, the vector need to have single acknowledgment locales for the regularly utilized restriction compounds as nearness of more than one acknowledgment destinations inside the vector will generate a few pieces, which will muddle the gene cloning. The ligation of unfamiliar DNA is done at a restriction site present in one of the two anti-microbial obstruction genes.
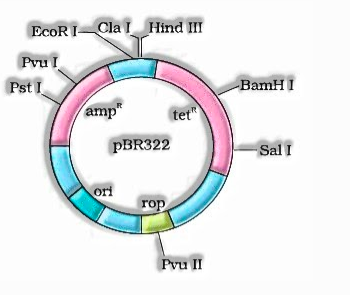
- coli cloning vector pBR322 indicating restriction destinations (Hind III, EcoR I, BamH I, Sal I, Pvu II, Pst I, Cla I), ori and anti-microbial opposition genes (ampR and tetR ). rop codes for the proteins engaged with the replication of the plasmid.
Insertional inactivation:
The most productive strategy for screening for the nearness of recombinant plasmids depends on the rule that the cloned DNA section disturbs the coding sequence of a gene. This is named as Insertional Inactiviation.
For instance, the ground-breaking technique for screening for the nearness of recombinant plasmids is alluded to as Blue-White determination. This technique depends on the insertional inactivation of the lac Z gene present on the vector. The lac Z gene encodes the chemical beta-galactosidase, which can separate a chromogenic substrate into a blue shaded item. On the off chance that this lac Z gene is inactivated by addition of an objective DNA section into it, the improvement of the blue shading will be forestalled and it gives white hued settlements. By along these lines, we can separate recombinant (white shading) and non-recombinant (blue shading) settlements.
- Vectors for cloning genes in plants and creatures Agrobacterium tumefacient (microorganism of dicot plant) can convey a bit of DNA known as ‘T-DNA” to change ordinary plant cells into a tumor and direct these tumor cells to deliver the synthetic compounds required by the microbe. Retroviruses in creatures can change typical cells into carcinogenic cells. The tumor inciting (Ti) plasmid of Agrobacterium tumefaciens has been altered into cloning vector having not any more pathogenic to plant. So also retrovirus have been adjusted into cloning vector for creatures.
Equipped host (For Transformation with Recombinant DNA)
1) Simple synthetic treatment with divalent calcium particles builds the productivity of host cells (through cell divider pores) to take up the rDNA plasmids.
2) rDNA can likewise be changed into host cell by brooding both on ice, trailed by setting them quickly at 42oC (Heat Shock), and afterward placing them back aside briefly. This empowers the microbes to take up the recombinant DNA.
3) In Microinjection strategy, rDNA is legitimately infused into the core of cells by using a glass micropipette.
4) Biolistics/Gene weapon strategy, it has been formed to bring rDNA into for the most part plant cells by using a Gene/Particle firearm. In this strategy, tiny particles of gold/tungsten are covered with the DNA of intrigue and shelled onto cells.
5) The last strategy utilizes “Incapacitated Pathogen” Vectors (Agrobacterium tumefaciens), which when permitted to contaminate the cell, move the recombinant DNA into the host.
Procedures of Recombinant DNA Technology
Recombinant DNA innovation includes a few stages in explicit sequence-
- Seclusion of DNA
- Discontinuity of DNA by restriction endonucleases
- Seclusion of an ideal DNA part
- Ligation of the DNA section into vector
- Changing the recombinant DNA into the host
- Refined the host cells in a medium everywhere scale
- Extraction of the ideal item.
- Isolation of Genetic material:Genetic material is detached from different macromolecules by using chemicals, for example, lysozyme (microscopic organisms), cellulase (plant cells), chitinase (growth). DNA that different out can be evacuated by spooling. The RNA can be evacuated by treatment with ribonuclease while proteins can be expelled by treatment with protease.
- Cutting of DNA at explicit area is performed by using restriction catalyst and Agarose gel electrophoresis to check the movement of a restriction compound absorption. Subsequent to cutting wellsprings of DNA just as vector DNA with a particular restriction compound to remove ‘gene of enthusiasm’ from the source DNA.
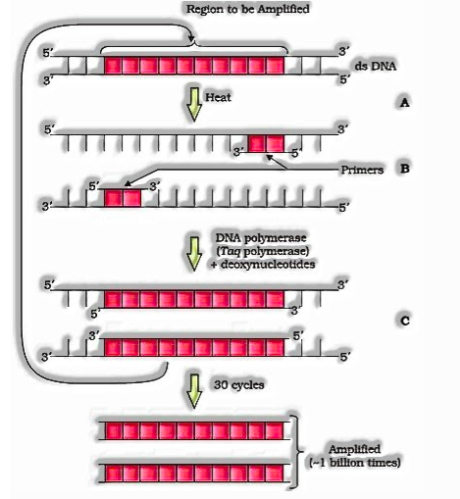
- Amplification of Gene of Interest using PCR( Polymerase Chain Reaction) to get different duplicates of the DNA or gene of enthusiasm for vitro by using set of groundworks and protein DNA polymerase.
Polymerase chain response (PCR) :
Each cycle has three stages: (A) Denaturation; (B) Primer strengthening; and (C) Extension of groundworks
This rehashed intensification is finished by the utilization of a thermostable DNA polymerase (segregated from a bacterium, Thermus aquaticus), which stay dynamic during the high temperature instigated denaturation of twofold abandoned DNA.
- Insertion of Recombinant DNA into the Host Cell/Organism incorporates making the beneficiary cells skilled to get, take up DNA present in its encompassing and so forth. The recombinant DNA bearing gene for protection from an anti-infection is moved into E.coli cells, the host cell become changed into ampicillin-obstruction cells.
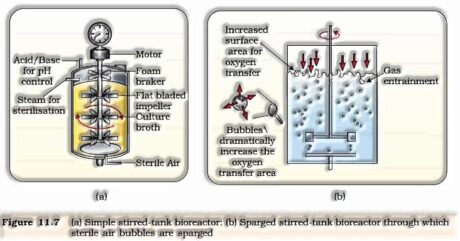
- Obtaining the unfamiliar gene item – the unfamiliar DNA increases in plant or creature cell to deliver attractive protein. Articulation of unfamiliar genes in host cells include, enhanced condition to acquire recombinant protein. The recombinant cell is duplicated in a nonstop culture framework wherein utilized medium is emptied out of one side while new medium is added from the other to keep up the cells in their physiological dynamic stage. A bioreactor gives the ideal conditions to accomplishing the ideal item by giving ideal development conditions (temperature, pH, substrate, salts, nutrients, oxygen).

- Downstream Processing includes forms that cause the item to acquire prepared for promoting. This procedure incorporates partition and purging called as downstream preparing. Appropriate additives are added to it and send for clinical preliminary if there should be an occurrence of medications before delivering to advertise for open use.
Question
Q1: Which of the accompanying sequences isn’t palindromic?
5’GGATCC 3′
3’CCTAGG 5′
5’AGCT 3′
3’TCGA 5′
5’TGCT 3′
3’TCGA 5′
5’TCGA 3′
3’AGCT 5′
Sol: The right answer is the choice ‘c’. The sequence on the two strands isn’t similar when both are perused in a similar direction. For example in 5′ to 3′ bearing or 3′ to 5′ heading.